A case study on Microbial Diversity of Bovine Mastitis Milk
Situation
Bovine mastitis is probably the most important disease for the dairy industry worldwide, causing economic losses due to reduced milk production, discarded milk, premature culling and excessive antibiotic use. It is also a serious bovine health issue associated with pain as well as reduced well-being of the affected animals [1]. This has become an alarming issue in certain parts of Sri Lanka due to lack of modern farming methods and proper hygienic practices. A preliminary survey on incidences of mastitis in Sri Lanka revealed an estimated loss of Rs. 4.3 million occurring annually due to this condition [2]. Bovine mastitis exists in unnoticeable subclinical form for months, spreading from one location to another, with high tendency of being nonresponsive to therapy. Chronically infected cows act as carriers and they impose a severe economic burden [3].
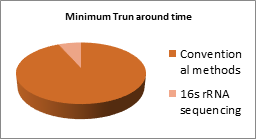
Bovine mastitis has a complex aetiology, being caused by a variety of bacteria and also by chemical and physiological factors. Streptococcus agalatiae is found to be the commonest agent causing chronic contagious mastitis. Staphylococcus aureus, S. dysgalatiae,S.uberis, Mycoplasma spp,Esherichia coli, environmental streptococci spp., Pseudomonas spp, yeast, fungi and alga such as Prototheca also cause disease implications [4]. Identification of bacteria causing this intramammary infection is the basis for controlling mastitis and targeting antimicrobial therapy. In the identification procedures, bacterial culture-based methods have long served as the gold standard for mastitis testing. However, these conventional tests are time consuming, with a routine turnaround time of 7 days. It also requires operator expertise, leading to differences in the results between different testing laboratories. Additionally, 25%-40% of milk samples from clinical and subclinical mastitis show no bacterial growth in conventional culturing. To overcome drawbacks of culture based techniques, real-time PCR was used for disease identification and pathogen classification with the aid of commercially available kits [6]. Although the technique could eliminate the shortcomings, disease detection using this technique required prior knowledge on the causal organism. Due to the complex aetiology of mastitis, however,prior knowledge on exact causal organism is rare. In addition, interpretation of results depends on a fluorescent labeling mechanism, which is relatively costlier compared to culture based techniques, in turn limiting the number of organisms which could be checked during a single run. Apart from real-time PCR, multiplex PCR (mPCR), denaturing gradient gel electrophoresis (DGGE) PCR, and PCR single-strand conformation polymorphism (SSCP) are now being used to identify bacterial DNA in milk samples. But these techniques hardly facilitate testing of bulk milk samples, which is a major drawback in large scale industrial testing.
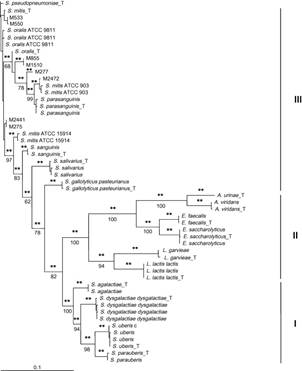
Fig- Phylogram of Streptococcus spp. and related bacteria isolated from bovine IMI associated with chronic mastitis[7]
 Solution
Solution
Sequencing and analysis of hypervariable regions within the 16S rRNA gene can provide relatively rapid and cost-effective, large scale methods for assessing bacterial diversity and abundance and is a definite advantage for pathogen discovery and identification. This technique uses raw milk as a starting material, does not require bacterial culturing, takes only 10 hours to complete the run, and enables a dramatic increase in throughput via parallel in depth analysis of many samples with lower costs. The analytical specificity and sensitivity of the test is unmatched and the test organisms can be classified up to species level while identifying their intragenic variations as well as relative abundances. Furthermore, the correlation of these bacteria to bovine chronic mastitis and their phylogeny can be investigated. Thus, 16s rRNA gene sequencing can be very efficient as a massively parallel technique that allows large scale testing of industrial raw milk samples. Thus, dairy processors can look forward to more detailed, cheaper, rapid and accurate identification of pathogens residing in their milk samples.
To summarize, 16s rRNA Sequencing has following advantages compared to other techniques-
| Conventional techniques | 16s rRNA Sequencing |
|---|---|
| Longer turnaround times. A single test requires a minimum of 7 days | Lesser turnaround times. Extremely rapid with a run time of only 12 hours. |
| Test accuracy is dependent on operator expertise. The accuracy of testing Is,̴80%[9] | Accuracy is operator-independent. Highly accurate with an accuracy of 99.9% |
| Fastidious, unculturable organisms and unusual biochemical reactions impede the identification process. | Test method is culture independent; Growth requirements, biochemical reactions and culture failures do not affect the identification process. |
| Only a limited number of organisms can be identified. Important organisms in a test sample can be missed out. | Identifies the complete range of organisms present in a test sample. |
| The percentage of strains correctly identified to the species level is sometimes less than satisfactory. | Permits identification of complete spectrum of organisms down to species level, their intragenic variations and relative abundances |
| Limited throughput, Sensitivity may vary. | Dramatic increase in throughput; High sensitivity |
References-
- Oikonomou, Georgios, et al. “Microbial diversity of bovine mastitic milk as described by pyrosequencing of metagenomic 16s rDNA.” PloS one10 (2012): e47671.
- Wickramasuriya, U. G. J. S. “EVALUATION OF A MASTITIS CONTROL PROGRAMME ADOPTED IN SMALL DAIRIES IN THE DISTRICT OF COLOMBO.” (1985).
- Thilakarathne D.S., Abeynayaken P et al “Bacterial Isolates and In Vitro Biogram of Milk Collected from Cows with Chronic Mastitis in the Central Province of Sri Lanka” Proceedings of the Peradeniya University Research Sessions, Sri Lanka, Vol. 16, 24th November 2011.
- Ravindran,V. Sriskandarajah, N. Rajamahendran, R.. 2010. Diseases and parasites of domesticated animals in Sri Lanka . [ONLINE] Available at: http://dl.nsf.ac.lk/handle/1/5159. [Accessed 25 September 13].
- ansci.illinois.edu. 2013. Mastitis Pathogens. [online] Available at: http://classes.ansci.illinois.edu/ansc438/mastitis/pathogens.html [Accessed: 22 Nov 2013].
- Koskinen, M. T., et al. “Field comparison of real-time polymerase chain reaction and bacterial culture for identification of bovine mastitis bacteria.”Journal of dairy science12 (2010): 5707-5715.
- Wyder, Anna Barbara, et al. “Streptococcus spp. and related bacteria: Their identification and their pathogenic potential for chronic mastitis–A molecular approach.” Research in veterinary science3 (2011): 349-357.
- Janda, J. Michael, and Sharon L. Abbott. “Bacterial identification for publication: when is enough enough?.” Journal of clinical microbiology6 (2002): 1887-1891.


